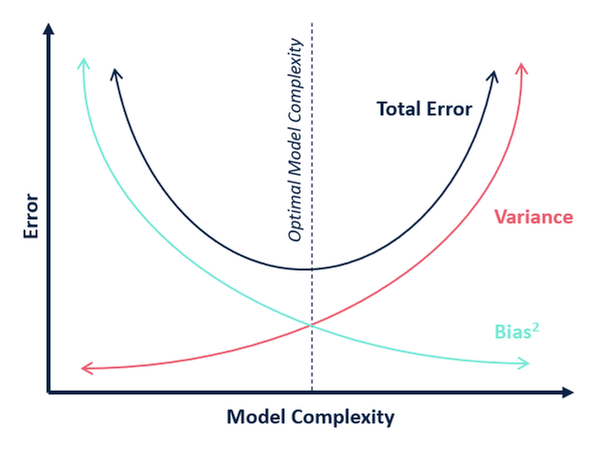
Bias-Variance Tradeoff¶
Bias and Variance Formulae¶
- Consider $y(x) = f^*(x) + \epsilon$
- $f^*$ is the true model
- $\epsilon$ is some $0$-mean noise with variance $\sigma^2$
- A model receives one dataset $S$ and outputs $\hat f$ as prediction of $y$. The error is:
$$\mathbb{E}[(y - \hat{f})^2] = \underbrace{{\sigma^2}}_\text{irreducible error} + \underbrace{{\text{Var}[\hat{f}]}}_\text{Variance} + \underbrace{(f - \mathbb{E}[\hat{f}])^2}_{\text{Bias}^2}$$
where expectation is taken w.r.t. all possible datasets, $\mathbb{E}[f(S)]=\mathbb{E}_S[f(S)]=\int f(S)p(S)dS$.
- Break error into two terms relating to $\mathbb{E}[\hat f]$ the "average" estimate over random datasets $S$.
- Irreducible error: Minimum possible error - determined by measurement noise
- Bias: Fitting capability - "complexity" of model
- Variance: Generalization capability - Model sensitivity to changes in dataset, (overfitting)
An example to explain Bias/Variance and illustrate the tradeoff¶
- Consider estimating a sinusoidal function.
(Example that follows is inspired by Yaser Abu-Mostafa's CS 156 Lecture titled "Bias-Variance Tradeoff")
Let's return to fitting polynomials¶
- Here we generate some samples $x,y$, with $y = \sin(\pi x)$
- We then fit a degree-$0$ polynomial (i.e. a constant function) to the samples
# polyfit_sin() generates 5 samples of the form (x,y) where y=sin(pi*x)
# then it tries to fit a degree=0 polynomial (i.e. a constant func.) to the data
# Ignore return values for now, we will return to these later
_, _, _, _ = polyfit_sin(degree=0, iterations=1, num_points=5, show=True)
We can do this over many datasets¶
- Let's sample a number of datasets
- How does the fitted polynomial change for different datasets?
# Estimate two points of sin(pi * x) with a constant 5 times
_, _, _, _ = polyfit_sin(degree=0, iterations=5)
What about over lots more datasets?¶
# Estimate two points of sin(pi * x) with a constant 100 times
_, _, _, _ = polyfit_sin(degree=0, iterations=100)
MSE, errs, mean_coeffs, coeffs_list = polyfit_sin(degree=0, iterations=100, num_points=3, show=False)
biases, variances = calculate_bias_variance(coeffs_list,RANGEXS,TRUEYS)
plot_bias_and_variance(biases,variances,RANGEXS,TRUEYS,np.polyval(np.poly1d(mean_coeffs), RANGEXS))
- Decomposition: $\mathbb{E}[(y - \hat{f})^2] = \underbrace{{\sigma^2}}_\text{irreducible error} + \underbrace{{\text{Var}[\hat{f}]}}_\text{Variance} + \underbrace{(f - \mathbb{E}[\hat{f}])^2}_{\text{Bias}^2}$
- Blue curve: true $f$
- Black curve: $\hat f$, average predicted values of $y$
- Yellow is error due to Bias, Red/Pink is error due to Variance
Bias vs. Variance¶
- We can calculate how much error we suffered due to bias and due to variance
poly_degree, results_list = 0, []
MSE, errs, mean_coeffs, coeffs_list = polyfit_sin(poly_degree, 500,num_points = 5,show=False)
biases, variances = calculate_bias_variance(coeffs_list,RANGEXS,TRUEYS)
sns.barplot(x='type', y='error',hue='poly_degree', data=pd.DataFrame([{'error':np.mean(biases), 'type':'bias','poly_degree':0},
{'error':np.mean(variances), 'type':'variance','poly_degree':0}]));
Let's now fit degree=3 polynomials¶
- Let's sample a dataset of 5 points and fit a cubic poly
MSE, _, _, _ = polyfit_sin(degree=3, iterations=1)
Let's now fit degree=3 polynomials¶
- What does this look like over 5 different datasets?
_, _, _, _ = polyfit_sin(degree=3,iterations=5,num_points=5,show=True)
Let's now fit degree=3 polynomials¶
- What does this look like over 50 different datasets?
# Estimate two points of sin(pi * x) with a line 50 times
_, _, _, _ = polyfit_sin(degree=3, iterations=50)
MSE, errs, mean_coeffs, coeffs_list = polyfit_sin(3,500,show=False)
biases, variances = calculate_bias_variance(coeffs_list,RANGEXS,TRUEYS)
plot_bias_and_variance(biases,variances,RANGEXS,TRUEYS,np.polyval(np.poly1d(mean_coeffs), RANGEXS))
$$\mathbb{E}[(y - \hat{f})^2] = \underbrace{{\sigma^2}}_\text{irreducible error} + \underbrace{{\text{Var}[\hat{f}]}}_\text{Variance} + \underbrace{(f - \mathbb{E}[\hat{f}])^2}_{\text{Bias}^2}$$
- Blue curve: true $f$
- Black curve: $\mathbb{E}[\hat f]$, average prediction of $y$
- Yellow is error due to Bias, Red/Pink is error due to Variance
Bias and Variance for different degree sizes¶
sns.barplot(x='type', y='error',hue='poly_degree',data=pd.DataFrame(results_list));
- High degree polys have lower bias but much greater variance!
- Bias-Variance Trade-off
Examples - Linear Regression¶
Consider $m$ features $\phi(x)$ and $n$ data points of noise level $\sigma^2$.
The trade-off is approximately (very crude!) $$ \mathbb{E}[(y - \hat{f})^2] = \underbrace{O({\sigma^2})}_\text{irreducible error} + \underbrace{O(\sigma^2\frac{m}{n})}_\text{Variance} + \underbrace{O(m^{-p})}_{\text{Bias}^2} $$
- Variance: $m$ competes with $n$.
- Bias reduces as $m$ increases.
Examples - Ridge Regression¶
Adding the regularization factor $\lambda$, the trade-off is modified with an effective number of features $$ \mathbb{E}[(y - \hat{f})^2] = \underbrace{O({\sigma^2})}_\text{irreducible error} + \underbrace{O(\sigma^2\frac{{\tilde{m}}}{n})}_\text{Variance} + \underbrace{O({\tilde{m}}^{-p})}_{\text{Bias}^2} $$ where $$ {\tilde{m}} = \sum_{j=1}^m \frac{\nu_i}{\nu_i+\lambda} $$ and $\nu_i$'s are the eigenvalues of $\Phi^\top\Phi$.
- For $\lambda>0$, ${\tilde{m}}<m$ - effectively reduces model complexity.
- When $\lambda=0$, ${\tilde{m}}=m$ and simple linear regression is recovered.
- When $\lambda\rightarrow\infty$, ${\tilde{m}}\rightarrow 0$ and we get a naive model (always predicts 0).
For more precise expressions and more challenging cases such as Lasso, see this note.
Methods for Model Selection¶
Choosing the model with the right complexity
- e.g. choose $\alpha$ in Ridge Regression, see [PRML] Chp. 3.5 for more details
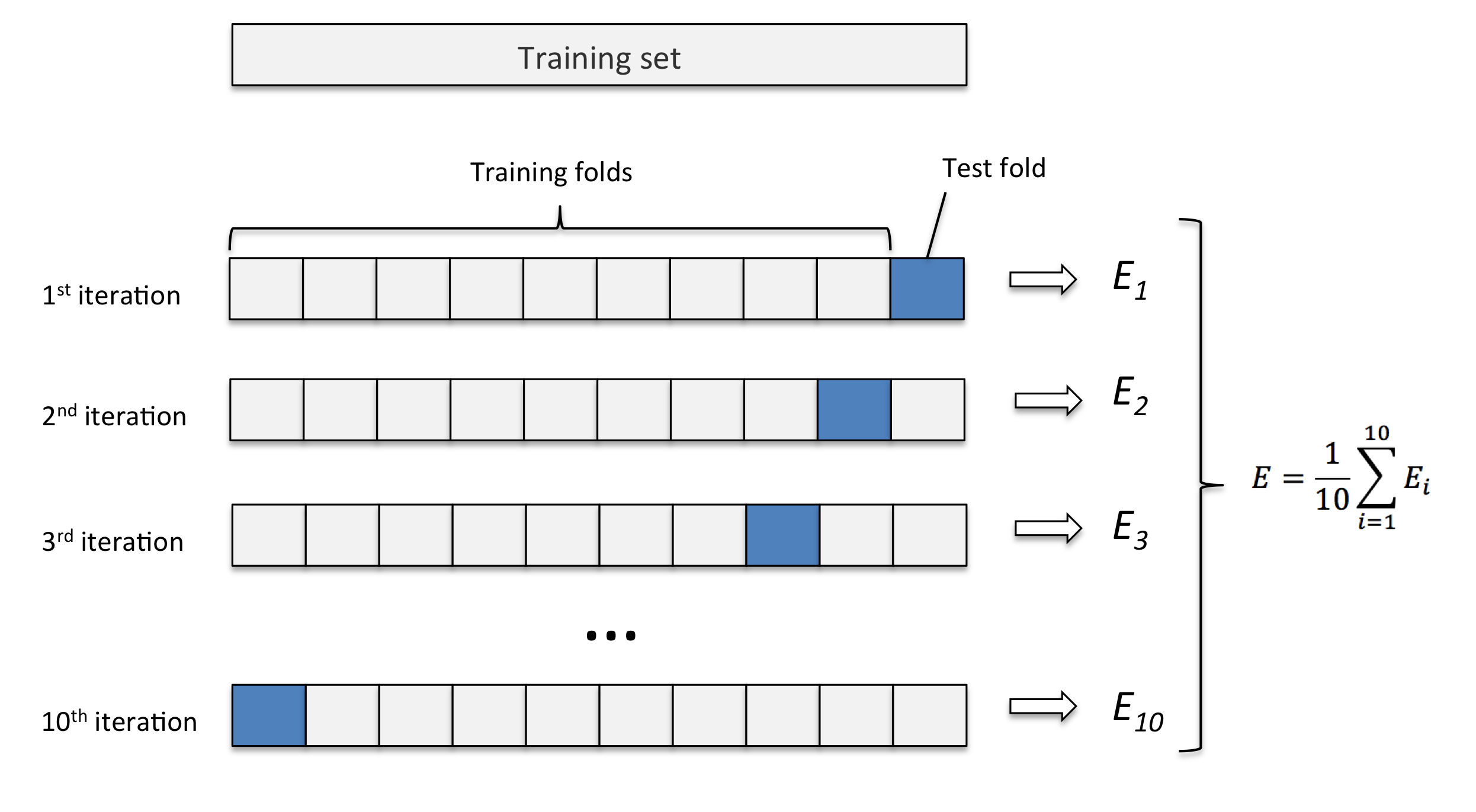
Cross-Validation¶
For a model $M(\alpha)$ with complexity parameter $\alpha$
- Split the dataset into K-fold
- For i=1,...,K
- Remove the ith fold and train a model
- Compute the error $E_i$ on the ith fold
- Average all the errors $E_1$,...,$E_K$ to obtain the estimated error of $M(\alpha)$
Repeat for different $\alpha$'s and find the one with minimum error
- Leave-One-Out (LOO): Divide a dataset having N samples into N-fold
- Advantages:
- Good when not many samples are available (otherwise one can create datasets for training and validation)
- Attenuates impact of overfitting - trying out many possible combinations of datasets
- Disadvantages:
- Computationally expensive - K-fold CV requires training the model K times
- Inconvenient if the model has multiple complexity parameters - Grid-search? Could be expensive
Information criteria¶
- Akaike information criteria (AIC) $$ AIC = 2M - 2\ln p(t|w_{ML}) $$
- Bayesian information criteria (BIC) $$ BIC = M\ln N - 2\ln p(t|w_{MAP}) $$
- $M$: number of complexity parameters, $N$: number of samples
- For linear regression: $$ \begin{aligned} -2\ln p(t|w) &= -N \ln\beta + N \ln{2\pi} + 2\beta E_D(w) \\ &= N\ln\frac{E_D(w)}{N} + const \end{aligned} $$
- Both are easy to evaluate - but not straightforward for interpretation
- BIC penalizes more on model complexity than AIC
- BIC works better when $N\gg M$
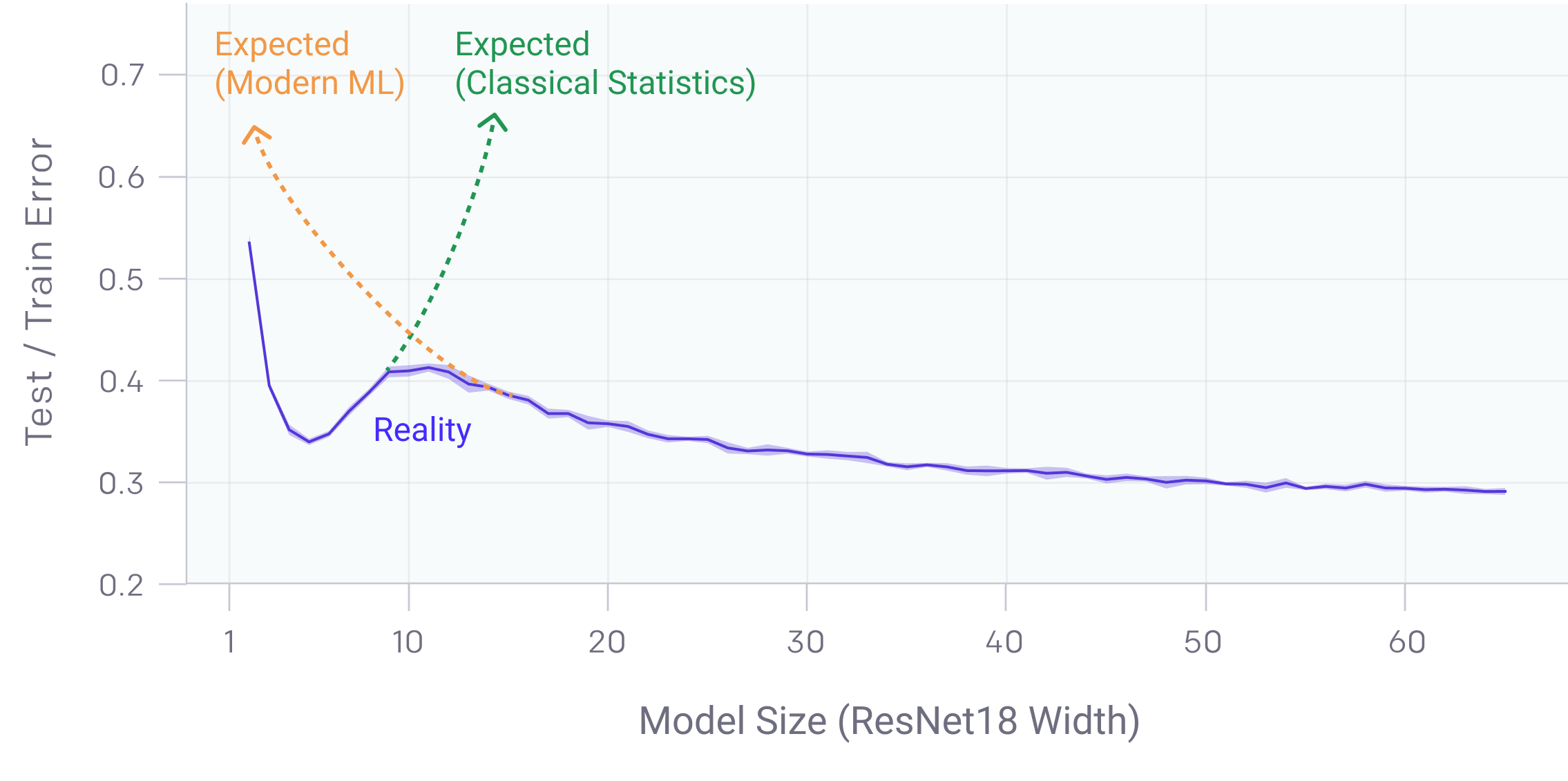
Deep Double Descent in Over-parametrized Case¶
AKA in deep learning: https://openai.com/blog/deep-double-descent/
A simple experiment: Consider a quantity $y$ and its measurements (i.e. features) $x_1,x_2,\cdots,x_F$
- The first $K$ features are $x_i=z_i+y$, where $z_i$ is unit Gaussian noise
- The latter $F-K$ features are merely noise, $x_i=z_i$
We will fit a series of models for $y$ using 100 samples with increasing number of features $M$.
- When $M\leq 100$, regular least squares suffices;
- When $M>100$ least norm squares is needed (but pseudo inverse just works fine).
Intuition:
- The first $K$ features carry information about $y$ - so the more of these features are used, the better is the prediction
- The rest $M-K$ features should not improve the prediction
But is this the case?
f = plt.figure(figsize=(6,4))
plt.plot(nins, ets, 'b-', label='Training')
plt.plot(nins, ess, 'r-', label='Test')
plt.plot([nfea, nfea], [0,1], 'k--')
plt.legend()
plt.ylim([0,1])
plt.xlabel('# of features')
plt.ylabel('Average error');
In fact, beyond the Bias-Variance curve, the test error still drops, after a short increase.
- For this particular problem, the extra features help cancel out the noise in the "useful" features.
- Still an open question for deep learning community...