TODAY: Quick (re)view of linear algebra¶
- Vectors and Matrices
- Eigenvalue decomposition
- Singular value decomposition (SVD)
References:¶
- [Kreyszig] Chap. 7-8
- [PRML] Appendix C
- Matrix Norms
Vector norms¶
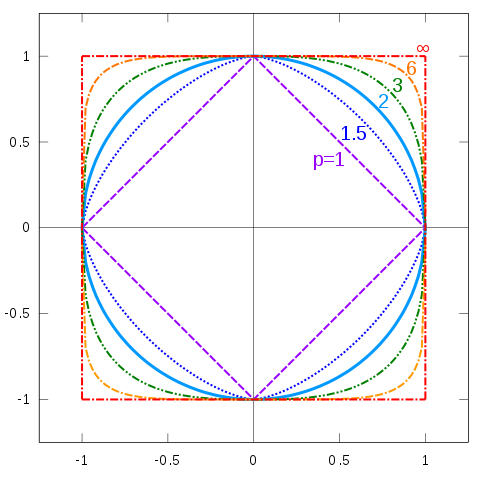
- Perhaps the most common norm is the Euclidean norm $$ \|x\|_2 := \sqrt{x_1^2 + x_2^2 + \ldots x_n^2}$$
- This is a special case of the $p$-norm: $$ \|x\|_p := \left(|x_1|^p + \ldots + |x_n|^p\right)^{1/p}$$
- Special cases: $p=1$ and $p=0$? $$ \|x\|_1\ \mbox{Sum of absolute values},\quad \|x\|_0\ \mbox{Number of non-zeros} $$
- There's also the so-called infinity norm $$ \|x\|_\infty := \max_{i=1,\ldots, n} |x_i|$$
- A vector $x$ is said to be normalized if $\|x\| = 1$
Geometrical interpretation¶
In 2D, when $\|x\|=1$, what is the shape of curve?
Matrix norms¶
Simple extension from vector norms - think a $m\times n$ matrix as a $mn$-dimensional vector
- For example, Frobenius norm $$ \|A\|_F := \left( \sum_{i,j} A_{ij}^2 \right)^{1/2} $$
- Induced norms from vector norms $$ \|A\|_p := \max_{\|x\|_p=1}\|Ax\|_p $$
Orthogonal + Normalized = Orthonormal¶
- Two vectors $x,y$ are orthogonal if $x^T y = 0$
- A square matrix $U \in \mathbb{R}^{n \times n}$ is orthogonal if all columns $u_1, \ldots, u_n$ are orthogonal to each other (i.e. $u_i^\top u_j = 0$ for $i \ne j$)
- $U$ is orthonormal if it is orthogonal and the columns are normalized, i.e. $\|u_i\|_2 = 1$ for every $i$.
- $U$ forms an orthonormal basis of the $\mathbb{R}^n$ vector space.
Orthonormal matrix for projection¶
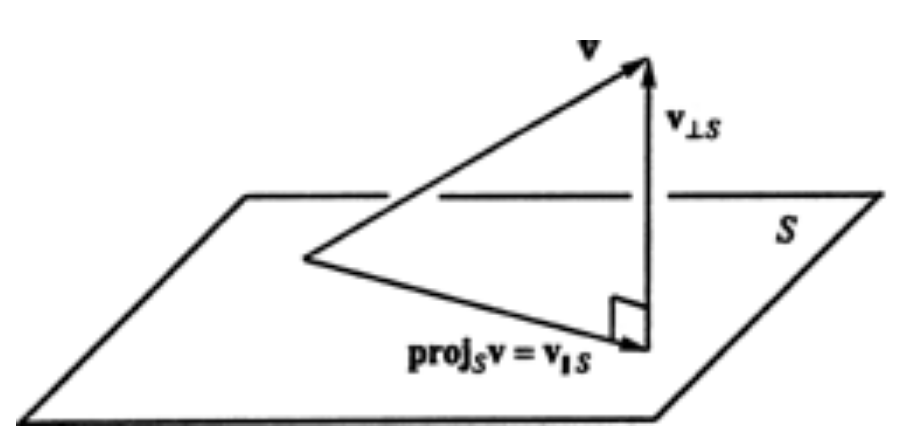
- $U\in \mathbb{R}^{n\times m}$ column-wise orthonormal, i.e. $U^\top U=I_m$
- Then $U$ form the orthonormal basis of a $m$-dimensional subspace $S$ in $\mathbb{R}^n$
- e.g. 2D plane in 3D space
- The component $p$ of a $n$-dimensional vector $v$ in subspace $S$ is given by the projection $ p=UU^\top v $
- The residual is $r = v-p = (I-UU^\top)v$ and orthogonal to $p$
- Or - the residual does not carry any information concerning the subspace
Positive Definiteness¶
- We say a symmetric matrix $A$ is positive definite if $$ x^\top A x > 0 \text{ for all } x \ne 0$$
- We say a matrix is positive semi-definite (PSD) if $$ x^\top A x \geq 0 \text{ for all } x$$
- A matrix that is positive definite gives us a norm. Let $$ \| x \|_A := \sqrt{x ^\top A x} $$
Visualization of PD matrices¶
Contour plots of $f(x)=x^TAx$, starting with -
$$ A=\begin{bmatrix} 1 & 0 \\ 0 & 4 \end{bmatrix} $$A = np.array([
[1,1],
[1,4]])
pltPSD(A, with_eig=False, if3d=False)
pltPSD(A, with_eig=False, if3d=True)
Eigenvalues and Eigenvectors¶
What are eigenvectors?¶
- A Matrix is a mathematical object that acts on a (column) vector, resulting in a new vector, i.e. $Ax=b$
- An eigenvector is the resulting vector that is parallel to $x$ (some multiple of $x$) $$ {A}x=\lambda x $$
- The eigenvectors with an eigenvalue of zero are the vectors in the nullspace
- If A is singular (takes some non-zero vector into 0) then $\lambda=0$
Numerical example¶
X = np.array([[3, 1], [1, 3]])
Matrix(X)
eigenvals, eigenvecs = np.linalg.eig(X)
Matrix(eigenvecs)
newX = eigenvecs.dot(np.diag(eigenvals)).dot(eigenvecs.T)
Matrix(newX)
Trace related to eigenvals¶
- Let $A$ be a matrix whose eigenvalues are $\lambda_1, \ldots, \lambda_n$
- Then we have the trace of satisfying $$\text{tr}(A) = \sum_{i=1}^n \lambda_i$$
X = np.random.randn(5,10)
A = X.dot(X.T) # For fun, let's look at A = X * X^T
eigenvals, eigvecs = np.linalg.eig(A) # Compute eigenvalues of A
sum_of_eigs = sum(eigenvals) # Sum the eigenvalues
trace_of_A = A.trace() # Look at the trace
(sum_of_eigs, trace_of_A) # Are they the same?
Determinant related to eigenvals¶
- Let $A$ be a matrix whose eigenvalues are $\lambda_1, \ldots, \lambda_n$
- Then we have the trace of satisfying $$|A| = \prod_{i=1}^n \lambda_i$$
# We'll use the same matrix A as before
prod_of_eigs = np.prod(eigenvals) # Sum the eigenvalues
determinant = np.linalg.det(A) # Look at the trace
(prod_of_eigs, determinant) # Are they the same?
Eigenvalue Decomposition¶
- Let $A$ be a matrix whose eigenvalues are $\lambda_1, \ldots, \lambda_n$ $$\Lambda=Diag([\lambda_1, \ldots, \lambda_n])$$
- And the corresponding eigenvectors are $x_1, \ldots, x_n$ $$X=[x_1, \ldots, x_n]$$
- Then we have the following decomposition $$ A=X\Lambda X^{-1} $$
- Further if $X$ is orthonormal, i.e. $X^{-1}=X^\top$, $$ A=\sum_{i=1}^n \lambda_i x_i x_i^\top $$
- Round back to visualization
A = np.array([
[1,1],
[1,2]])
pltPSD(A, with_eig=True, if3d=False)
Singular Value Decomposition¶
- Not all square matrices can do eigenvalue decomposition - but
- Any matrix (symmetric, non-symmetric, etc.) $A \in \mathbb{R}^{n\times m}$ admits a singular value decomposition (SVD)
- The decomposition has three factors, $U \in \mathbb{C}^{n \times n}$, $\Sigma \in \mathbb{R}^{n \times m}$, and $V \in \mathbb{C}^{m \times m}$ $$A = U \Sigma V^*$$
- $U$ and $V$ are both orthonormal matrices, and $\Sigma$ is diagonal. $V^*$ indicates conjugate transpose.
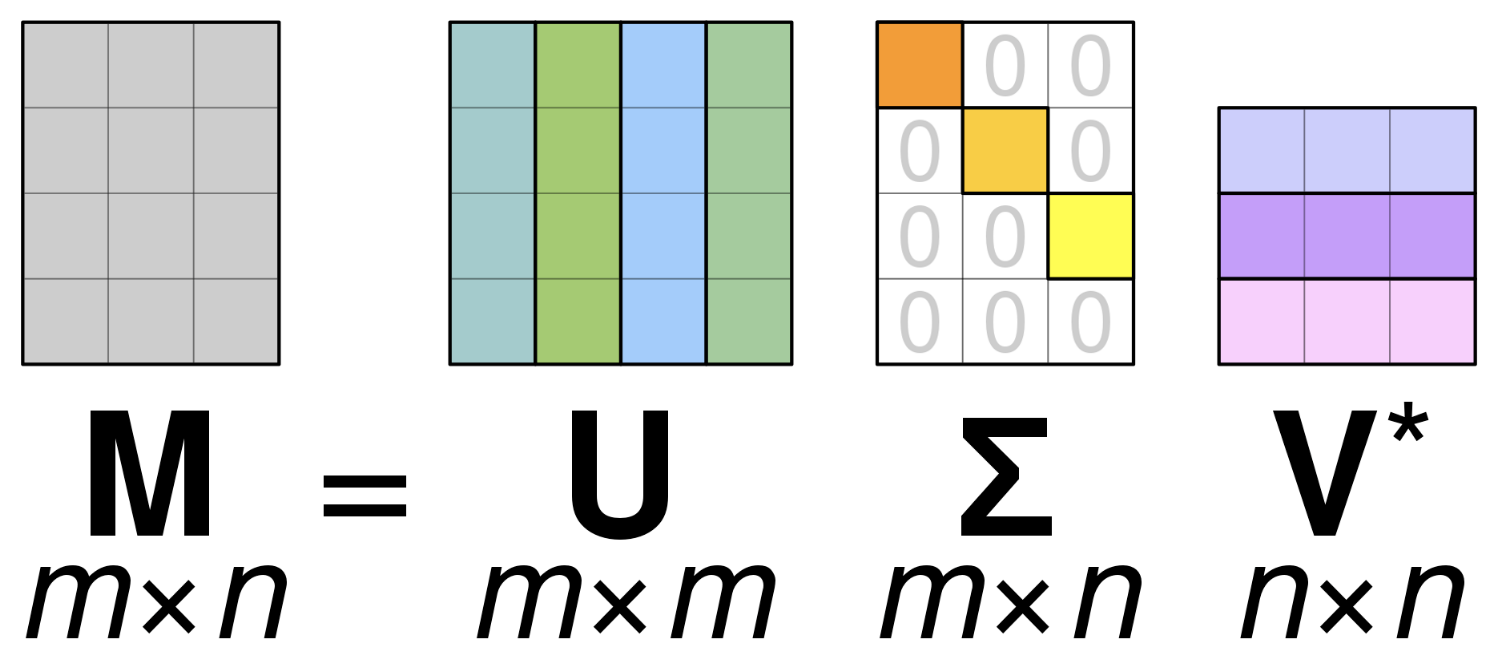
SVD Example¶
A = np.array([[4, 4], [-3, 3]])
Matrix(A)
- Let's show Sigma from the SVD output
U, Sigma_diags, V = np.linalg.svd(A)
Matrix(np.diag(Sigma_diags)) # Numpy's SVD only returns diagonals, here I'm showing full Sigma
- And we can show the orthonormal bases $U$ and $V$
Matrix(U), Matrix(V) # I rounded the values for clarity
Properties of the SVD¶
For simplicity, let's assume everything is real; then conjugate transpose is just transpose.
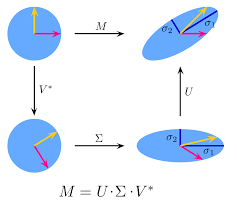
$$\text{SVD:} \quad \quad A = U \Sigma V^\top$$- The singular values of $A$ are the diagonal elements of $\Sigma$
- The singular values of $A$ are the square roots of the eigenvalues of both $A^\top A$ and $A A^\top$
- The left-singular vectors of $A$, i.e. the columns of $U$, are the eigenvectors of $A A^\top$
- The right-singular vectors of $A$, i.e. the columns of $V$, are the eigenvectors of $A^\top A$
Also - $\|A\|_2 = \sigma_\max(A)$, $\|A\|_F = (\sum_{i=1}^n \sigma_i^2)^{1/2}$
- An analogy to eigenvalue decomposition - but applicable to all matrices $$ A=\sum_{i=1}^{n} \sigma_iu_iv_i^\top $$
- Common trick to truncate and approximate a matrix - say the last $m$ $\sigma$'s are close to zero
- Error estimate
- What is the implication in (1) subspace projection and (2) data compression?
Simple application of SVD: Condition number¶
Consider two sets of linear equations $$ \begin{bmatrix} 3 & 1 \\ -3.0001 & 1 \end{bmatrix} \begin{bmatrix} x_1 \\ x_2 \end{bmatrix} = \begin{bmatrix} 4 \\ 4.0001 \end{bmatrix} $$
$$ \begin{bmatrix} 3 & 1 \\ -2.9999 & 1 \end{bmatrix} \begin{bmatrix} x_1 \\ x_2 \end{bmatrix} = \begin{bmatrix} 4 \\ 4.0002 \end{bmatrix} $$How different are their solutions?
A1=np.array([
[3,1],
[-3.0001,1]])
b1=np.array([4,4.0001])
A2=np.array([
[3,1],
[-2.9999,1]])
b2=np.array([4,4.0002])
x1=np.linalg.solve(A1,b1)
x2=np.linalg.solve(A2,b2)
print(x1)
print(x2)
[-1.66663889e-05 4.00005000e+00] [-3.33338889e-05 4.00010000e+00]
Consider another two sets of linear equations - note the signs $$ \begin{bmatrix} 3 & 1 \\ 3.0001 & 1 \end{bmatrix} \begin{bmatrix} x_1 \\ x_2 \end{bmatrix} = \begin{bmatrix} 4 \\ 4.0001 \end{bmatrix} $$
$$ \begin{bmatrix} 3 & 1 \\ 2.9999 & 1 \end{bmatrix} \begin{bmatrix} x_1 \\ x_2 \end{bmatrix} = \begin{bmatrix} 4 \\ 4.0002 \end{bmatrix} $$How different are their solutions?
B1=np.array([
[3,1],
[3.0001,1]])
b1=np.array([4,4.0001])
B2=np.array([
[3,1],
[2.9999,1]])
b2=np.array([4,4.0002])
y1=np.linalg.solve(B1,b1)
y2=np.linalg.solve(B2,b2)
print(y1)
print(y2)
[1. 1.] [-2. 10.]
Condition number of a matrix, $\kappa(A)$: How much the solution $x$ would change with respect to a change in $b$.
$$ \kappa(A) = \|A\|\|A^{-1}\| = \frac{\sigma_\max(A)}{\sigma_\min(A)} $$print(np.linalg.cond(A1))
print(np.linalg.cond(B1))
3.0000500009374806 200006.00009591616
Special algorithms are needed to solve linear systems with ill-conditioned matrices!
- We will cover some of these algorithms later (LU, Cholesky, and QR decomposition)